At ISDC 2018, we gave the Dana Meadows Award for best student paper to Gizem Aktas, for Modeling the Biological Mechanisms that Determine the Dynamics of Stress Response of the Human Body (with Yaman Barlas). This is a very interesting paper that elegantly synthesizes literature on stress, mood, and hormone interactions. I plan to write more about it later, but for the moment, here’s the model for your exploration.
The dynamic stress response of the human body to stressors is produced by nonlinear interactions among its physiological sub-systems. The evolutionary function of the response is to enable the body to cope with stress. However, depending on the intensity and frequency of the stressors, the mechanism may lose its function and the body can go into a pathological state. Three subsystems of the body play the most essential role in the stress response: endocrine, immune and neural systems. We constructed a simulation model of these three systems to imitate the stress response under different types of stress stimuli. Cortisol, glucocorticoid receptors, proinflammatory cytokines, serotonin, and serotonin receptors are the main variables of the model. Using both qualitative and quantitative physiological data, the model is structurally and behaviorally well-validated. In subsequent scenario runs, we have successfully replicated the development of major depression in the body. More interestingly, the model can present quantitative representation of some very well acknowledged qualitative hypotheses about the stress response of the body. This is a novel quantitative step towards the comprehension of stress response in relation with other disorders, and it provides us with a tool to design and test treatment methods.
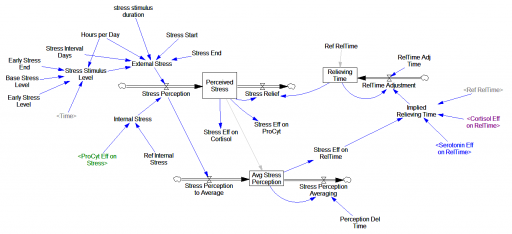
The original is a STELLA model; here I’ve translated it to Vensim and made some convenience upgrades. I used the forthcoming XMILE translation in Vensim to open the model. You get an ugly diagram (due to platform differences and XMILE’s lack of support for flow-clouds), but it’s functional enough to browse. I cleaned up the diagrams and moved them into multiple views to take better advantage of Vensim’s visual approach.
The model ran right away, though I had to add one MAX statement to handle a uniflow (not supported in Vensim, and something I remain allergic to). There’s actually an important lesson on model replication and calibration in this.
When I first translated the model, I ran a few scenarios, using the comprehensive replication instructions in the supplemental material for the paper. I built up a Vensim command script to make it easy to replicate all the scenarios in the paper. To do that, I had to modify the equations a bit, so that manual equation editing (in STELLA) could be replaced by automatic parameter changes.
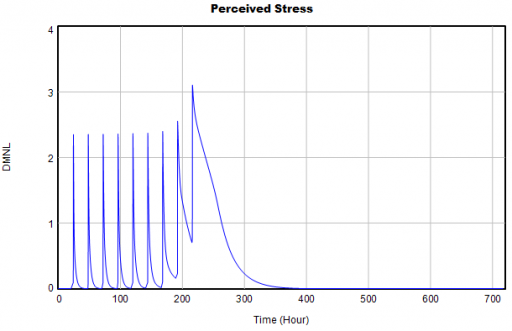
Then I ran my script and eyeballed a few graphs. Things looked pretty good:
The same, right? Not so fast! If you look closely, you’ll find that the Vensim version (bottom) has 9 peaks instead of 10, due to my replacement of a cascade of IF … ELSE test inputs with a simpler PULSE TRAIN. When you fix the count, there are still issues, because the duration parameter for each pulse (0.2) is not an integral multiple of the TIME STEP. (Incidentally, differences arising from PULSE implementations are tricky – see Yutaka Takahashi’s poster from ISDC 2018).
It took me several iterations to work out what was going wrong. I found that, to really verify that the translation (plus my initially erroneous upgrades) was OK, I had to export a run from STELLA, import it as a dataset in Vensim, and compare behavior hour by hour. That’s how I discovered the subtle but important uniflow difference.
The fact that tiny differences in test input implementations matter highlights the extreme numerical sensitivity of the model. This is a feature, not a bug. It arises from positive feedback that creates sensitive thresholds in stress response: 5% more episodic stress can be the difference between routine recovery and total collapse.
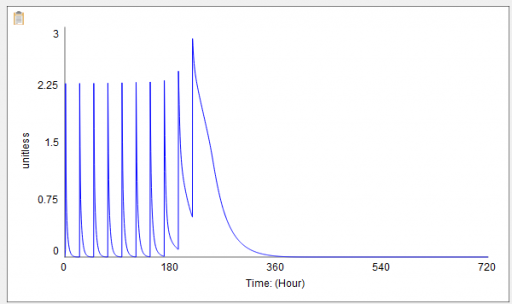
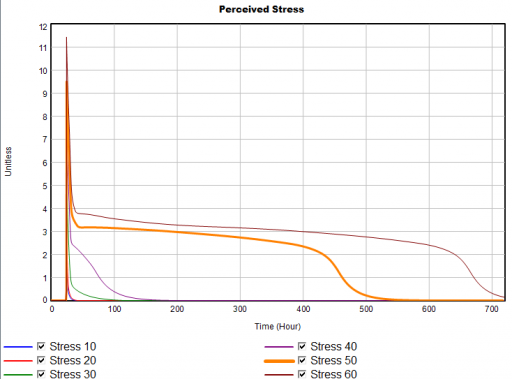
For example, here’s a sensitivity experiment with external stress at 10, 20, 30, 40, 50 & 60 units:
Notice that for external stress <= 40, recovery is quick – hours to days. But somewhere above 40 is a nonlinear threshold, beyond which recovery takes weeks.
This .zip archive contains:
- An updated source model (.stmx) from the author, used for the translation.
- The translated model (.mdl and .vpm). This version won’t work in PLE because it uses macros, but you can use the free Model Reader to run it.
- Command scripts for replicating the paper’s scenarios, plus the vector of stress levels above.
StressResponseModel_converted 7.zip
Update: StressResponseModel_converted 7b.zip fixes a unit error in a test input (my mistake) – this version is closest to the original in the paper.
Update 2: StressResponseModel_converted 8.zip has an improved control panel and runs 4x faster. It departs from the original to improve sensitivity analysis capability and pulse test stability, but remains dynamically identical (as far as I can determine).
The original paper and supplementary material should be in the conference submission system.
Stay tuned for more on this topic! Here’s a detailed critique & analysis.
Hi Tom and friends,
Thanks for refering to my paper.
Each software has its own implementation.
Users need to check if their intents are expressed correctrly.
But, I found many SD software users skip it.
Undoubtly, using something correctly is essential for getting something meaningful.
However, some software is too “friendly” to check something deeply.
We always need to check if our software expresses our models precisely, even if the software reports no errors!